Argument parsers statistics
Common Workflow Language command line tools mostly consist of input/output parameters descriptions. These descriptions could be created automatically and directly from the code of the actual Python tool. The more information is provided in the source code of the tool, the less has to be written by hand in a CWL tool description.
The amount of information about tool arguments is defined by its argument parser. Different argument parsers have different syntax and verbosity level, that’s why it is important to know:
a) which argument parsers are used most frequently;
b) which ones provide the most information that could be used in forming CWL command line tools.
After a brief overview I have selected 5 most popular ways to parse command line arguments in Python:
sys.argv
Standard module sys provides the most straightforward way to parse command line arguments, simply putting all of them into a list, regardless of their characteristics. This is the least flexible way to deal with arguments, but due to its simplicity and universality, it is very popular
argparse
This is a standard module in Python since version 2.7. It has a lot of advantages over its predecessor optparse, such as handling positional arguments, supporting sub-commands, producing more informative usage messages. For now, it is the most convenient way to obtain information about parameters and to form CWL tool out of it.
optparse
A deprecated but still used module in old projects.
docopt
A library for “creating beautiful command-line interfaces”, as claimed by its developers. It uses just a few lines of code to generate a parser and requires a well-written docstring.
click
A library which is considered by some more intuitive, lightweight than argparse and at least as powerful.
I was curious about which libraries are the most wide-spread in the bioinformatics community. That’s why I wrote a simple program to gather some statistic from GitHub repositories. I parsed a list of bioinformatics repositories from Python Package Index, grabbed links to GitHub sources and counted a number of files where each of the above libraries is imported. The source code is available here.
The results are the following:
| optparse | 130 files |
| docopt | 47 files |
| argparse | 727 files |
| argv | 804 files |
| click | 85 files |
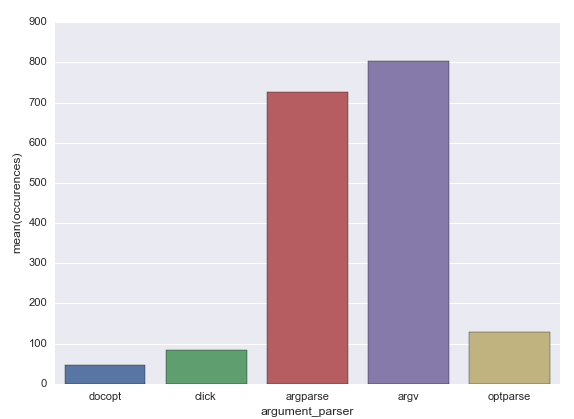
So, the most popular argument parsers are the standard libraries: sys.argv and argparse exceed other libraries by number at least by five times.
It was also interesting to look at argument parsers in the context of the popularity of the repositories. Here is the plot which represents occurrences of argument parsers in different repositories regarding the number of their downloads over the last month, a dot means the presence of a library in a repository.
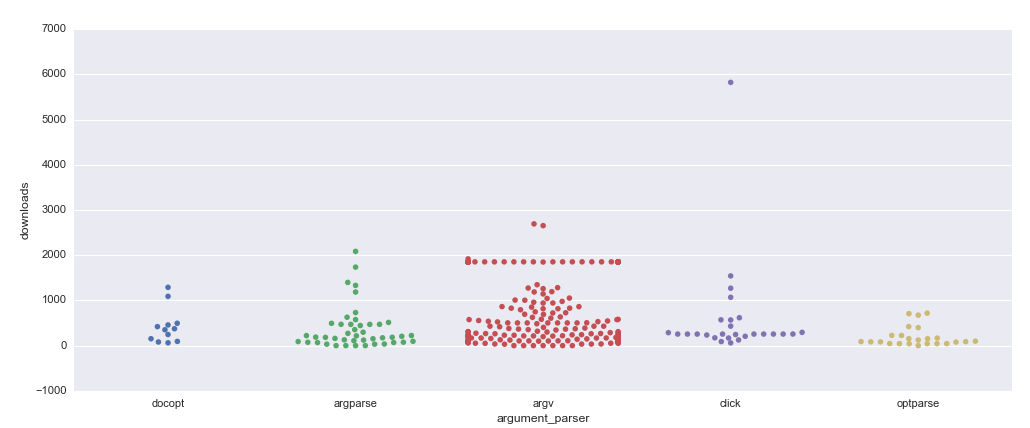 There are much more
There are much more sys.argv dots than argparse dots, though the number of files is pretty close; it means that projects that don’t use argument parsers intensively, one or two times per project, tend to use sys.argv. It is also more widespread among popular repositories, but I looked through some of them, and they tend to use argument parsers in targets others than bio tools which I am supposed to convert.
A useful outcome for the project: top 10 repositories which use argparse.
| Name | Description | Argparse | Downloads |
| resolwe-bio | Bioinformatics pipelines for the Resolwe platform. | 37 | 722 |
| pyfastaq | Script to manipulate FASTA and FASTQ files, plus API for developers | 37 | 1043 |
| lisa | 3D data processing toolbox | 36 | 1069 |
| svtools | Tools for processing and analyzing structural variants | 27 | 230 |
| pyGenClean | Automated data clean up pipeline for genetic data | 26 | 353 |
| phylotoast | Useful additions to the QIIME analysis pipeline including tools for data visualization and cluster-computing. | 25 | 98 |
| peyotl | Library for interacting with Open Tree of Life resources | 23 | 31 |
| plastid | Convert genomic datatypes into Pythonic objects useful to the SciPy stack | 18 | 287 |
| taxtastic | Tools for taxonomic naming and annotation | 17 | 267 |
| loom-engine | loom workflow engine | 15 | 53 |
In general, the statistics shows that argparse2cwl project has good chances to be useful for a wide circle of cases. Even though the most popular argument parser is not appropriate for processing, there are more than enough tools which use argparse. Nevertheless, the number of usages of other argument parsers is not so insignificant to neglect them, so it would be nice to add their support too.